Clinical Interpretation of genetic variants by ACMG/AMP 2015 guideline
InterVar is a bioinformatics software tool for clinical interpretation of genetic variants by the ACMG/AMP 2015 guideline. The input to InterVar is an annotated file generated from ANNOVAR, while the output of InterVar is the classification of variants into 'Benign', 'Likely benign', 'Uncertain significance', 'Likely pathogenic' and 'Pathogenic', together with detailed evidence code.
The American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published in 2015 the updated standards and guidelines for the clinical interpretation of sequence variants with respect to human diseases, based on 28 criteria. However, variability between individual interpreters may be extensive due to reasons such as the different understanding of these guidelines and the lack of standard algorithms to implement these guidelines, yet computational tools for semi-automated variant interpretation are not available. To address these problems, we proposed a suite of methods to implement these criteria, and developed a tool called InterVar to facilitate human reviewers to interpret the clinical significance of variants. InterVar can take a pre-annotated file or a VCF file as input, and generate automated interpretation on 18 criteria. Furthermore, we developed a companion web server wInterVar to enable user-friendly variants interpretation with an automated interpretation step and a manual adjustment step. These tools are especially useful to address severe congenital or very early onset developmental disorders with high penetrance. Using results from a few published sequencing studies, we demonstrated the utility of InterVar in significantly reducing the time to interpret clinical significance of sequence variants.
Figure 1 .The flowchart of the two-step procedure of InterVar. We used underlined and bold fonts to denote automated criteria.(click to enlarge)
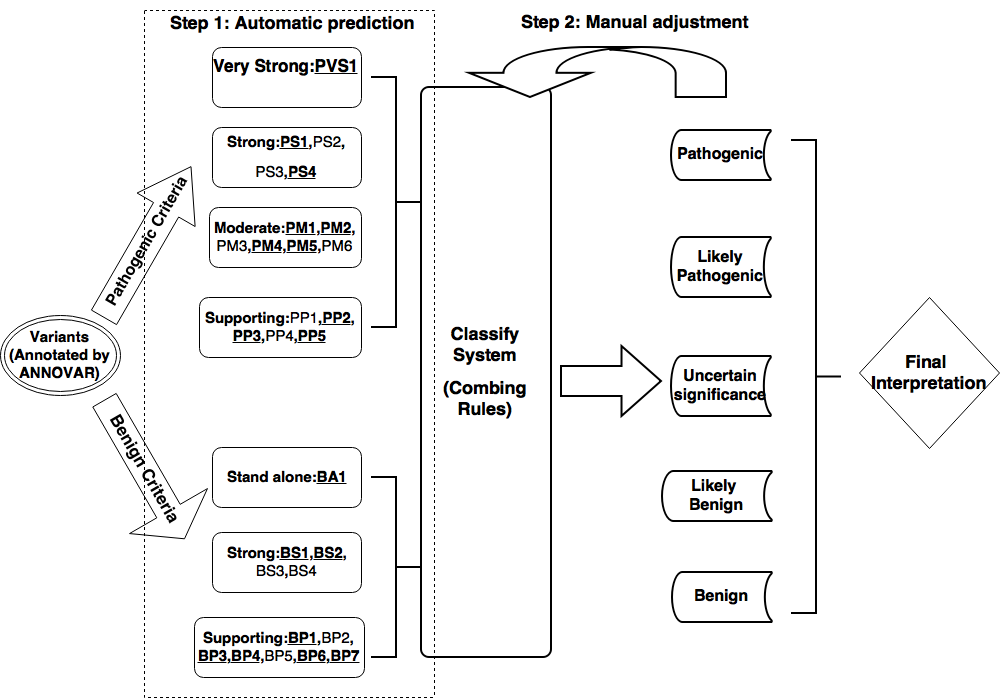
Figure 2 .An illustration of the 28 criteria from the ACMG/AMP2015 guidelines. For some criteria, we denote the name of internal database and their size within the parenthesis.(click to enlarge)
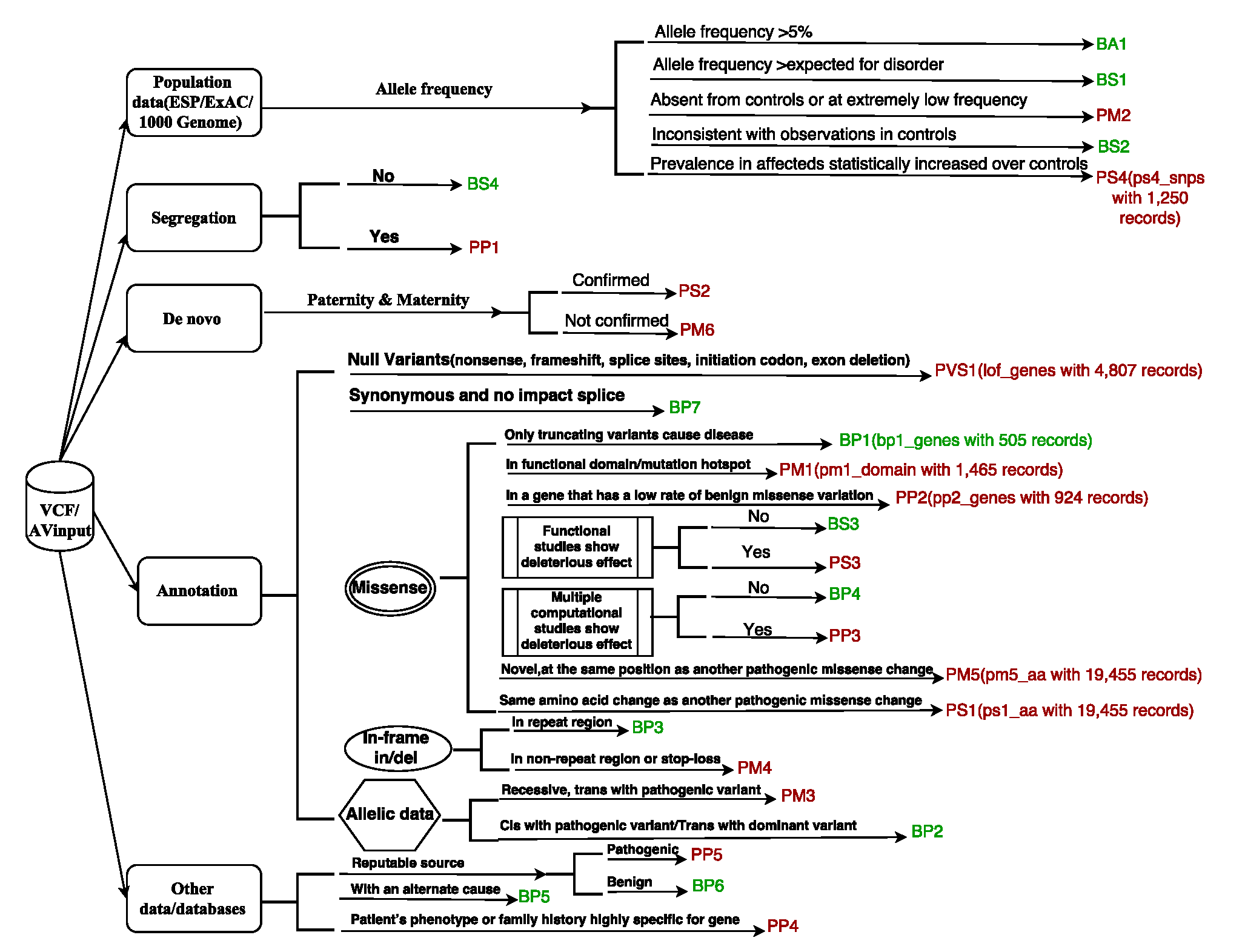
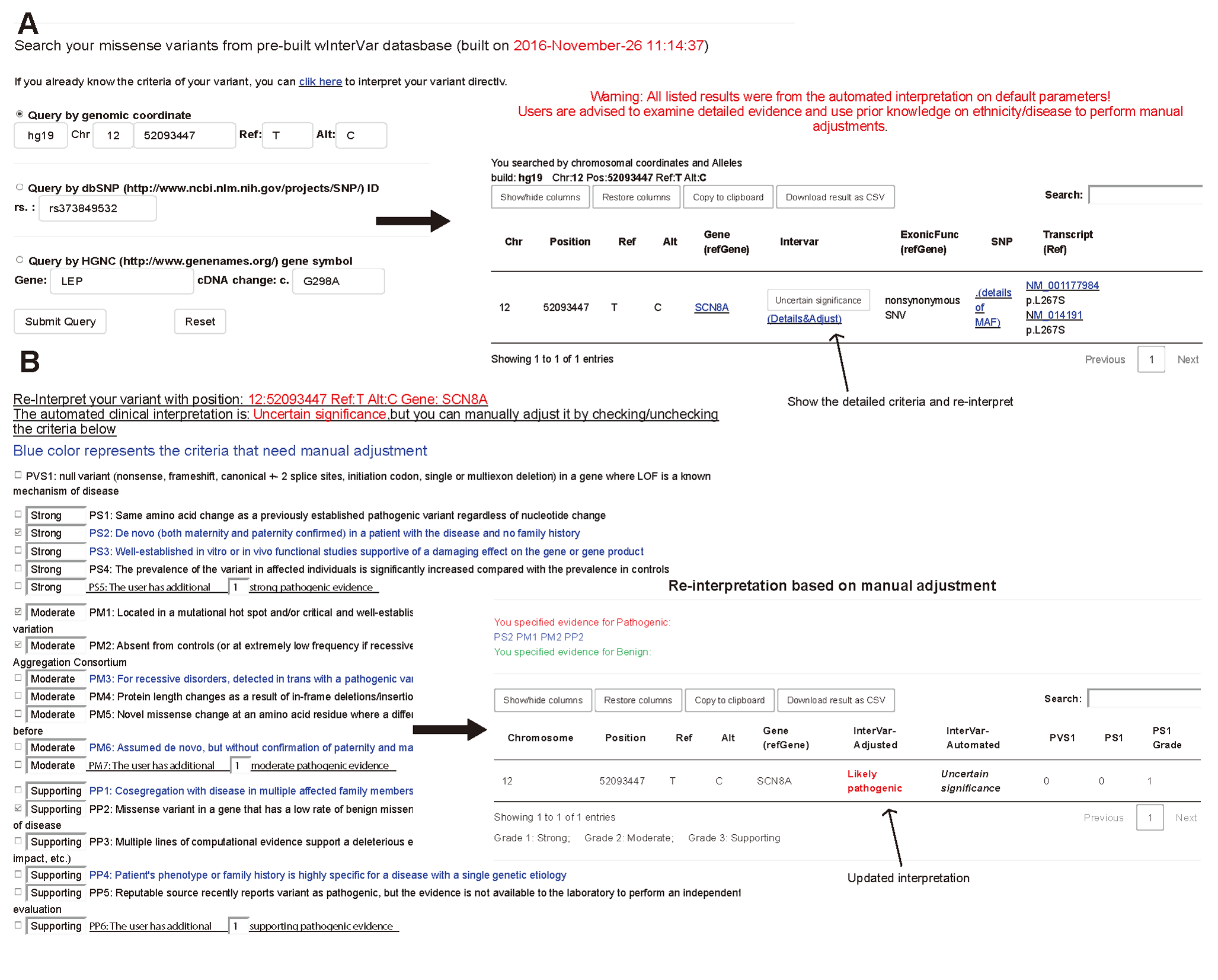
Figure 3. An illustration of wInterVar. (A) automatic interpretation of genetic variants, which can be entered by several means (B) once users click "Adjust", the full list of criteria is shown for manual adjustment, to derive the final results.
Supplementary Materials:
1. LOF genes list for PVS1
2. Rare and Non-conflicting exonic variants submitted by MedGen in Clinvar-2016-0302